5 Linear Models
5.2 Sums of Squares Types
Getting Started
To demonstrate the various types of sums of squares, we’ll create a data frame called df_disease taken from the SAS documentation (reference). The summary of the data is shown.
## # A tibble: 3 x 3
## drug disease y
## <fct> <fct> <dbl>
## 1 1 1 42
## 2 1 1 44
## 3 1 1 36## drug disease y
## 1:18 1:24 Min. :-6.00
## 2:18 2:24 1st Qu.: 9.00
## 3:18 3:24 Median :21.00
## 4:18 Mean :18.88
## 3rd Qu.:28.00
## Max. :44.00
## NA's :14The Model
For this example, we’re testing for a significant difference in stem_length using ANOVA. In R, we’re using lm() to run the ANOVA, and then using broom::glance() and broom::tidy() to view the results in a table format.
lm_model <- lm(y ~ drug + disease + drug*disease, df_disease)The glance function gives us a summary of the model diagnostic values.
lm_model %>% glance()## # A tibble: 1 x 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.456 0.326 10.5 3.51 0.00130 11 -212. 450. 477.
## # ... with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>The tidy function gives a summary of the model results.
lm_model %>% tidy()## # A tibble: 12 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 29.3 4.29 6.84 0.0000000160
## 2 drug2 -1.33 6.36 -0.210 0.835
## 3 drug3 -13.0 7.43 -1.75 0.0869
## 4 drug4 -15.7 6.36 -2.47 0.0172
## 5 disease2 -1.08 6.78 -0.160 0.874
## 6 disease3 -8.93 6.36 -1.40 0.167
## 7 drug2:disease2 6.58 9.78 0.673 0.504
## 8 drug3:disease2 -10.9 10.2 -1.06 0.295
## 9 drug4:disease2 0.317 9.30 0.0340 0.973
## 10 drug2:disease3 -0.900 9.00 -0.100 0.921
## 11 drug3:disease3 1.10 10.2 0.107 0.915
## 12 drug4:disease3 9.53 9.20 1.04 0.306The Results
You’ll see that R print the individual results for each level of the drug and disease interaction. We can get the combined F table in R using the anova() function on the model object.
lm_model %>%
anova() %>%
tidy() %>%
kable()| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| drug | 3 | 3133.2385 | 1044.4128 | 9.455761 | 0.0000558 |
| disease | 2 | 418.8337 | 209.4169 | 1.895990 | 0.1617201 |
| drug:disease | 6 | 707.2663 | 117.8777 | 1.067225 | 0.3958458 |
| Residuals | 46 | 5080.8167 | 110.4525 | NA | NA |
And with some extra work, we can get a Total row to match the F table output by SAS.
lm_model %>%
anova() %>%
tidy() %>%
add_row(term = "Total", df = sum(.$df), sumsq = sum(.$sumsq)) %>%
kable()| term | df | sumsq | meansq | statistic | p.value |
|---|---|---|---|---|---|
| drug | 3 | 3133.2385 | 1044.4128 | 9.455761 | 0.0000558 |
| disease | 2 | 418.8337 | 209.4169 | 1.895990 | 0.1617201 |
| drug:disease | 6 | 707.2663 | 117.8777 | 1.067225 | 0.3958458 |
| Residuals | 46 | 5080.8167 | 110.4525 | NA | NA |
| Total | 57 | 9340.1552 | NA | NA | NA |
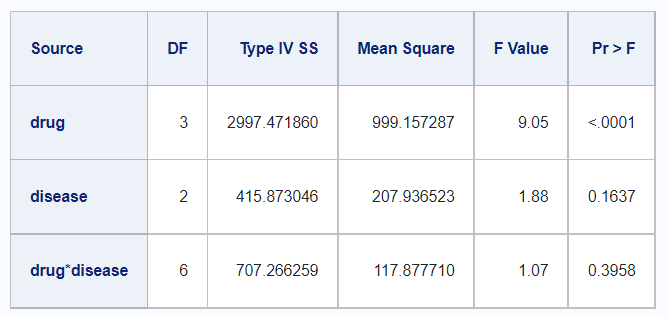
Comparing this to the output in SAS, we see the following.
proc glm;
class drug disease;
model y=drug disease drug*disease / ss1 ss2 ss3 ss4;
run;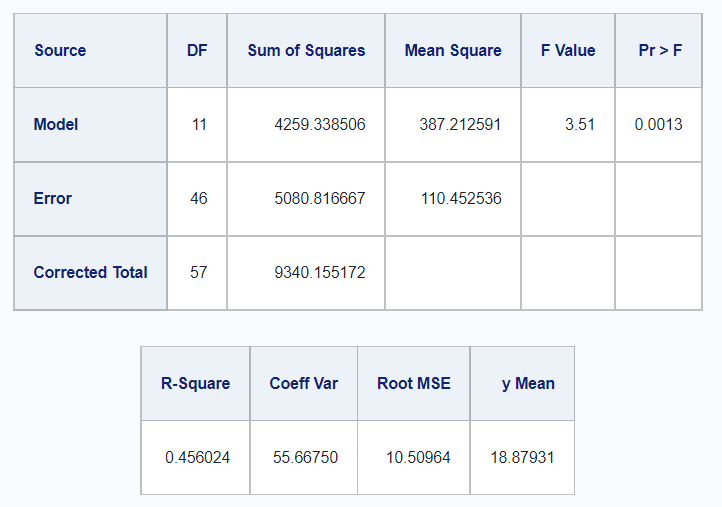
Sums of Squares Tables
In SAS, it is easy to find the tables for the variouse types of sums of squares calculations. Unfortunately, it is not easy to match this output in using functions from base R. However, the rstatix package offers a solution to produce these various sums of squares tables. Note that there does not appear to be a Type IV SS equivalent in R.
5.2.0.1 Type I
In R,
df_disease %>%
rstatix::anova_test(
y ~ drug + disease + drug*disease,
type = 1,
detailed = TRUE) %>%
rstatix::get_anova_table() %>%
kable()| Effect | DFn | DFd | SSn | SSd | F | p | p<.05 | ges |
|---|---|---|---|---|---|---|---|---|
| drug | 3 | 46 | 3133.239 | 5080.817 | 9.456 | 5.58e-05 | * | 0.381 |
| disease | 2 | 46 | 418.834 | 5080.817 | 1.896 | 1.62e-01 | 0.076 | |
| drug:disease | 6 | 46 | 707.266 | 5080.817 | 1.067 | 3.96e-01 | 0.122 |
And in SAS,
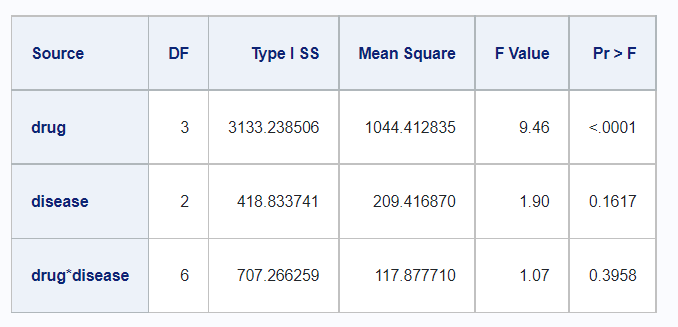
Type II
In R,
df_disease %>%
rstatix::anova_test(
y ~ drug + disease + drug*disease,
type = 2,
detailed = TRUE) %>%
rstatix::get_anova_table() %>%
kable()| Effect | SSn | SSd | DFn | DFd | F | p | p<.05 | ges |
|---|---|---|---|---|---|---|---|---|
| drug | 3063.433 | 5080.817 | 3 | 46 | 9.245 | 6.75e-05 | * | 0.376 |
| disease | 418.834 | 5080.817 | 2 | 46 | 1.896 | 1.62e-01 | 0.076 | |
| drug:disease | 707.266 | 5080.817 | 6 | 46 | 1.067 | 3.96e-01 | 0.122 |
And in SAS,
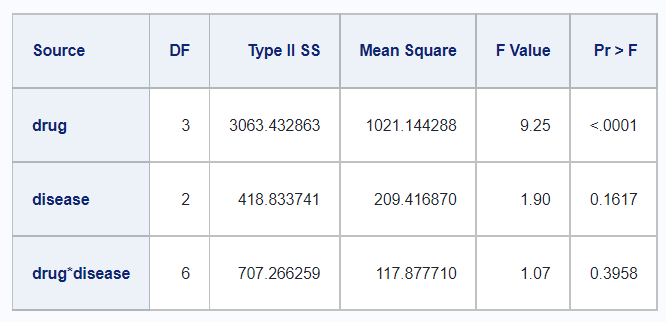
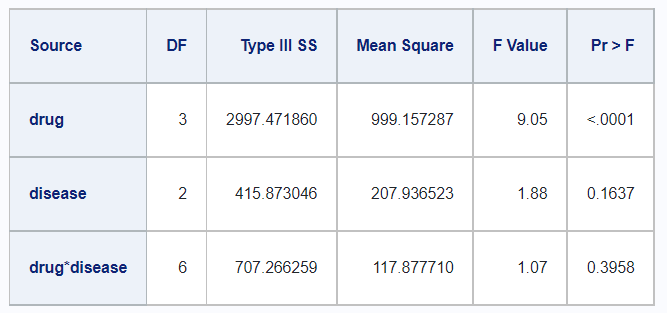
Type III
In R,
df_disease %>%
rstatix::anova_test(
y ~ drug + disease + drug*disease,
type = 3,
detailed = TRUE) %>%
rstatix::get_anova_table() %>%
kable()| Effect | SSn | SSd | DFn | DFd | F | p | p<.05 | ges |
|---|---|---|---|---|---|---|---|---|
| (Intercept) | 20037.613 | 5080.817 | 1 | 46 | 181.414 | 0.00e+00 | * | 0.798 |
| drug | 2997.472 | 5080.817 | 3 | 46 | 9.046 | 8.09e-05 | * | 0.371 |
| disease | 415.873 | 5080.817 | 2 | 46 | 1.883 | 1.64e-01 | 0.076 | |
| drug:disease | 707.266 | 5080.817 | 6 | 46 | 1.067 | 3.96e-01 | 0.122 |
And in SAS,
5.3 Contrasts
Getting Started
To demonstrate contrasts, we’ll create a data frame called df_trial. We see that the drug variable has three levels, A, C and E.
## drug pre post sex
## A:10 Min. : 3.00 Min. : 0.00 M:15
## C:10 1st Qu.: 7.00 1st Qu.: 2.00 F:15
## E:10 Median :10.50 Median : 7.00
## Mean :10.73 Mean : 7.90
## 3rd Qu.:13.75 3rd Qu.:12.75
## Max. :21.00 Max. :23.00## Rows: 30
## Columns: 4
## $ drug <fct> A, A, A, A, A, A, A, A, A, A, C, C, C, C, C, C, C, C, C, C, E, E,~
## $ pre <dbl> 11, 8, 5, 14, 19, 6, 10, 6, 11, 3, 6, 6, 7, 8, 18, 8, 19, 8, 5, 1~
## $ post <dbl> 6, 0, 2, 8, 11, 4, 13, 1, 8, 0, 0, 2, 3, 1, 18, 4, 14, 9, 1, 9, 1~
## $ sex <fct> M, F, M, F, M, F, M, F, M, F, M, F, M, F, M, F, M, F, M, F, M, F,~In order to work with these levels as contrasts, we can use one of the pre-existing R functions to create the identity matrix for this factor variable. The contr.treatment definition is the default one that R uses, while the contr.SAS is the definition that SAS uses by default.
contr.treatment(levels_drug)## C E
## A 0 0
## C 1 0
## E 0 1
contr.SAS(levels_drug)## A C
## A 1 0
## C 0 1
## E 0 0R also has the following definitions ready for use: contr.sum, contr.poly, and contr.helmert.
Using Contrasts in R
There are many ways to work with contrasts in R, but here we’re only going to focus on defining contrasts in the modeling function. In this example, we’re creating a linear model to predict post values from the pre and drug values. The only difference between the two is in the contrasts argument. In one case, we’re using the R default, and in the other we’re using the SAS default.
model_trt <-
lm(post ~ pre + drug, data = df_trial,
contrasts = list(drug = contr.treatment))
model_sas <-
lm(post ~ pre + drug, data = df_trial,
contrasts = list(drug = contr.SAS))Comparing the output for these two models, we see that differ in which drug level is being used as the reference baseline.
tidy(model_trt)## # A tibble: 4 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -3.88 1.99 -1.95 0.0616
## 2 pre 0.987 0.164 6.00 0.00000245
## 3 drug2 0.109 1.80 0.0607 0.952
## 4 drug3 3.45 1.89 1.83 0.0793
tidy(model_sas)## # A tibble: 4 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -0.435 2.47 -0.176 0.862
## 2 pre 0.987 0.164 6.00 0.00000245
## 3 drug1 -3.45 1.89 -1.83 0.0793
## 4 drug2 -3.34 1.85 -1.80 0.0835We can define a custom contrast as well. In this case, we are comparing A to E, where E is the baseline group. As is typical of contrasts, the values must sum to 0, where negative numbers represent the baseline groups.
It is possible to combine contrasts using cbind. Notice here that contrast definitions don’t require level names to be assigned, as long as the order of the levels is maintained.
Easy Contrasts in R with emmeans
The emmeans package makes working with contrasts much easier. In fact, this is the method that we recommend when trying to match contrast output between SAS and R.
We begin by defining some models.
model_lm <- lm(post ~ pre + drug + sex, data = df_trial)
model_av <- aov(post ~ pre + drug + sex, data = df_trial)
model_gm <- glm(post ~ pre + drug + sex, data = df_trial)Then we convert these models to emmeans models. In the emmeans function we can specify which variables we want to display estimated marginal means for.
model_lm %>% emmeans(specs = "drug")
model_av %>% emmeans(specs = "drug", by = "sex")
model_gm %>% emmeans(specs = ~ drug | sex)Or, we can define some common contrasts with the contrast function.
# All Pairwise Comparisons
model_lm %>%
emmeans("drug") %>%
contrast(method = "pairwise")
# Treatment v Control Comparison
model_gm %>%
emmeans("drug") %>%
contrast(method = "trt.vs.ctrl") We can also control the reference group, or reverse the contrast order using the arguments in the contrast function.
model_lm %>%
emmeans("drug") %>%
contrast(method = "trt.vs.ctrl", ref = 2)
model_lm %>%
emmeans("drug") %>%
contrast(method = "trt.vs.ctrl", rev = T) Custom contrasts can be defined as well.
Matching Contrasts: R and SAS
It is recommended to use the emmeans package when attempting to match contrasts between R and SAS. In SAS, all contrasts must be manually defined, whereas in R, we have many ways to use pre-existing contrast definitions. The emmeans package makes simplifies this process, and provides syntax that is similar to the syntax of SAS.
This is how we would define a contrast in SAS.
# In SAS
proc glm data=work.mycsv;
class drug;
model post = drug pre / solution;
estimate 'C vs A' drug -1 1 0;
estimate 'E vs CA' drug -1 -1 2;
run;And this is how we would define the same contrast in R, using the emmeans package.
lm(formula = post ~ pre + drug, data = df_trial) %>%
emmeans("drug") %>%
contrast(method = list(
"C vs A" = c(-1, 1, 0),
"E vs CA" = c(-1, -1, 2)
))Note, however, that there are some cases where the scale of the parameter estimates between SAS and R is off, though the test statistics and p-values are identical. In these cases, we can adjust the SAS code to include a divisor. As far as we can tell, this difference only occurs when using the predefined Base R contrast methods like contr.helmert.
proc glm data=work.mycsv;
class drug;
model post = drug pre / solution;
estimate 'C vs A' drug -1 1 0 / divisor = 2;
estimate 'E vs CA' drug -1 -1 2 / divisor = 6;
run;